Novel Design Strategy for Therapeutic Antisense
Oligonucleotides for Rare Genetic Disorders
Novel Design Strategy for Therapeutic Antisense
Oligonucleotides for Rare Genetic Disorders
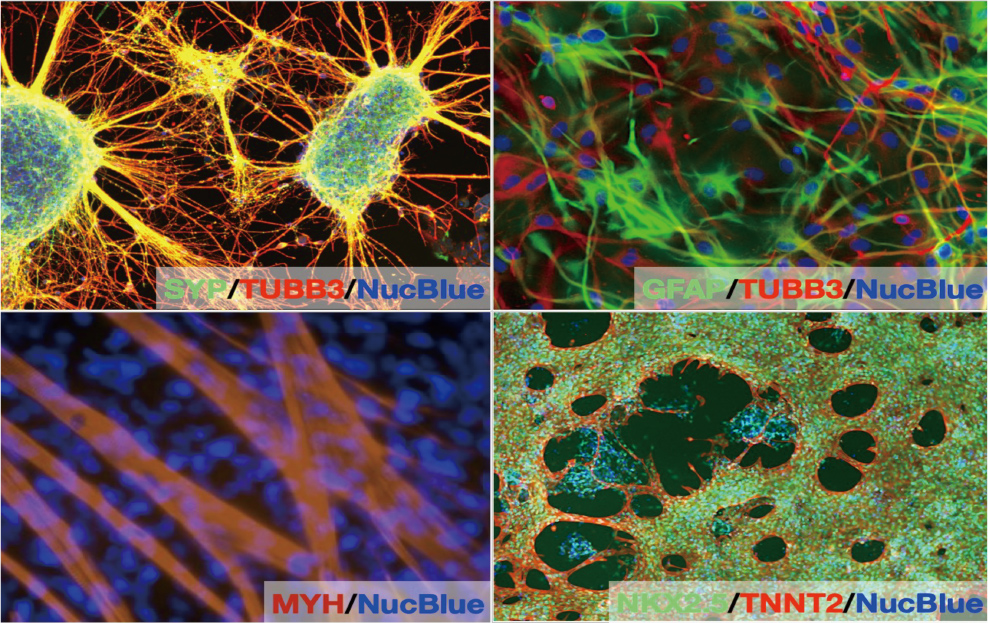
Genetic disorders are caused by genetic variations. Some genetic abnormalities can be improved to properly function, by removing exons or unwanted sequences from mRNA, such as exon skipping therapy for Duchenne muscular dystrophy. However, the search for target sequences requires a lot of money, time, and labor. We have developed a method to rationally design antisense oligonucleotides by targeting specific sequences. We aim to provide therapeutics for currently untreatable diseases such as muscular dystrophies, cystic fibrosis, familial colorectal adenomatosis, and others.
- Affiliation:
- Kyoto University
Graduate School of Medicine
- Representative:
- Tomonari Awaya

Kyoto University
Graduate School of Medicine
Associate Professor
Tomonari Awaya MD, PhD
He is a pediatrician, pediatric neurologist and clinical geneticist specializing in rare genetic disorders. He moved to a drug discovery laboratory targeting RNA splicing, where he is working on diagnosis of rare genetic disorders and their potential therapeutic intervention.
SOCIALSocial Implementation
Seeking a cure for rare genetic disorders with effective new drugs
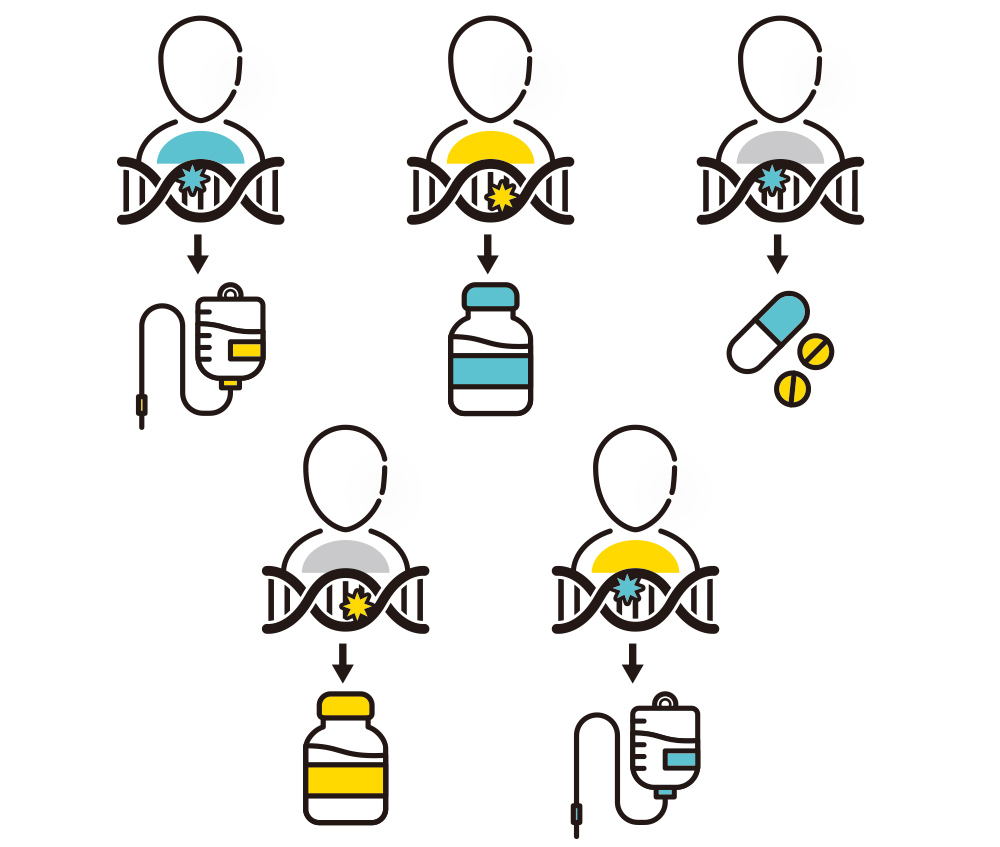
Many children with rare genetic disorders are faced with difficult circumstances. Since the diseases themselves are rare, there are limited resources and funding to develop clinical medications, and even though they appear to be effective, it is difficult to bring them into clinical practice. We started this project with the hope of eliminating these difficulties. We are also aiming for realization of personalized medicine in coordination with regulatory authorities so that effective drugs should be delivered even to a small number of patients, though clinical trials are quite difficult when targeting patients with rare disorders. Antisense oligonucleotides can be a good candidate to realize personalized medicine.
ORIGINALITYUniqueness, Passion for Development
We can streamline the search for target sequences to significantly reduce cost and labor
Until now, the search for the target sequence needed to skip a certain exon has required enormous cost and effort. In this research, we developed a method to rationally determine the target sequence, which significantly reduces the time, cost, and effort required. This allows us to efficiently test antisense oligonucleotides against many target exons, expanding the potential for many patients with rare genetic disorders amenable to exon-skipping therapies.

image photo
VISIONDream, Outlook, Business Image
We aim to realize personalized medicine through genetic disorders caused by RNA splicing abnormalities
It is possible to conduct clinical trials for specific genetic variants that cluster in certain ethnic groups within the current regulations. However, most genetic variants are usually sporadic, and such variant-specific drugs are difficult to be approved, though it is estimated that genetic variants that result in splicing alterations account for more than 15% of all variants. If our antisense oligonucleotide designing strategy works, we can reduce costs and effort and significantly accelerate the development process, thus expanding the potential for personalized medicine for rare genetic disorders.